23 minute read · November 26, 2025
Using Dremio, lakeFS & Python for Multimodal Data Management
· Head of DevRel, Dremio
· Product Manager, LakeFS

Multimodal data, ranging from images and logs to structured tables, powers today’s AI systems, but managing these diverse assets in sync remains a significant challenge. While software teams benefit from Git workflows to track code versions, data teams often lack equivalent tools to version, isolate, and merge changes across multiple data types. That’s where lakeFS steps in. By bringing Git-style version control to your data lake, lakeFS lets you branch and commit data changes safely. And when paired with Apache Iceberg for structured tables and Dremio for unified querying, you get a complete, AI-ready lakehouse experience. In this post, we’ll show how to build a multimodal lakehouse using lakeFS, Iceberg, Dremio, and Python, demonstrating how these tools work together to keep your data versioned, queryable, and production-ready.
What is lakeFS?
lakeFS is a control plane for AI-ready data, powered by a scalable data version control system that enables data and AI teams to manage data the way software engineers manage code. Using a Git-like model - branching, committing, and merging - lakeFS brings software engineering best practices into data, AI, and ML workflows. The result is faster, more reliable delivery of AI-ready data.
Key benefits of lakeFS include:
- Faster AI delivery: Safely collaborate and iterate on large, multimodal datasets without duplicating storage.
- Improved data reliability: Enforce data quality checks and catch issues early, preventing them from impacting production.
- Reproducible AI/ML results: Data versioning ensures training and experiments are repeatable and you can trace exactly which inputs produced each model.
- Reduced data access friction: Keep GPUs busy without waiting for data and manage all access from a single place.
lakeFS runs on top of object stores such as Amazon S3, Azure Blob Storage, Google Cloud Storage, and any S3-compatible system, such as MinIO, and integrates smoothly with any tool that interacts with the object store.
Try Dremio’s Interactive Demo
Explore this interactive demo and see how Dremio's Intelligent Lakehouse enables Agentic AI
Why Multimodal Data Is Hard to Manage - and How lakeFS Helps
Modern AI and ML workflows rely on multimodal data - a mix of structured data (like Iceberg tables), semi-structured (e.g., JSON or logs) and unstructured data (such as images, videos, or model artifacts). These pieces live in different systems, evolve at various speeds, and depend on one another. The hard part is keeping them in sync and under control while preserving the reproducibility of the exact data versions used.
lakeFS simplifies this by providing a unified versioning layer for all of it:
- Unstructured and semi-structured data via the object store API
- Structured data through a dedicated Iceberg REST catalog
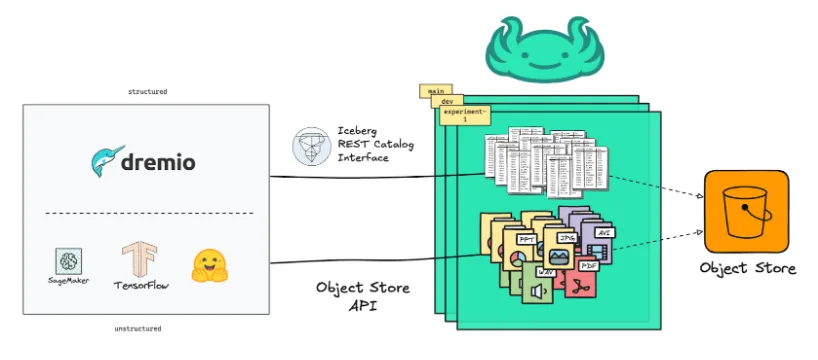
This unified approach ensures consistent version control, and governance across all data types, making multimodal data management straightforward. It enables teams to isolate changes with zero-copy branches, commit multimodal updates together, and merge once validated.
Using lakeFS with Iceberg?
lakeFS implements a spec-compliant Iceberg REST Catalog so that Dremio can connect to it as a standard REST catalog source. The catalog is version-aware: namespaces encode the lakeFS repository and reference (branch, commit, or tag), so every query is pinned to an exact snapshot.
The distinctive value of the lakeFS REST catalog lies in its multimodal data management. A single lakeFS snapshot captures both your Iceberg tables and the unstructured/semi-structured objects they depend on (images, models, logs), keeping tables and files aligned to the same version and ensuring reproducibility and traceability to multimodal changes.
The catalog also supports atomic commits across multiple tables, aligning with emerging work on multi-table transactions in the Iceberg ecosystem.
Ingesting Multimodal Data to lakeFS
To illustrate multimodal workflows on lakeFS, we use the PD12M dataset (images + metadata). Images are registered with lakeFS via zero-copy Import, and metadata is written to an Iceberg table via the lakeFS Iceberg REST Catalog. The result is a unified environment where tables and files share the same version.
Create a repository
We created a repo called multimodal-pd12m and a working branch, ingest, to add data in isolation.

Ingest the dataset images
We used Import from S3 to register all PD12M images under images/ in the repo; this was a metadata-only operation (no data copied).

Ingest the metadata
We downloaded the dataset’s Parquet metadata files, loaded them, and performed a small transformation: replacing the url attribute with a relative path pointing to the image in the lakeFS repo. For example, the url https://pd12m.s3.us-west-2.amazonaws.com/images/02fe431e-10d5-5416-afdd-c6811343222e.jpeg transformed to the path images/02fe431e-10d5-5416-afdd-c6811343222e.jpeg so that it is logical to reference dataset images using their lakeFS locations.
Write the Iceberg table via the lakeFS REST Catalog
Starting from a pandas DataFrame, we write the metadata as an Iceberg table using the lakeFS Iceberg REST Catalog, under multimodal-pd12m.ingest.images.image_metadata.”
from pyiceberg.catalog.rest import RestCatalog
import pandas as pd
catalog = RestCatalog(name="my_catalog", **{
'prefix': 'lakefs',
'uri': f'{lakefs_endpoint}/iceberg/api',
'oauth2-server-uri': f'{lakefs_endpoint}/iceberg/api/v1/oauth/tokens',
'credential': f'{lakefs_access_key}:{lakefs_secret_key}',
})
tbl_data = pa.Table.from_pandas(metadata_df)
table = catalog.create_table("multimodal-pd12m.ingest.images.image_metadata", schema=tbl_data.schema)
table.append(tbl_data)Publish the dataset
We merged the ingest branch into main and created a tag, baseline, to freeze the initial ingestion state even as main continues to evolve.
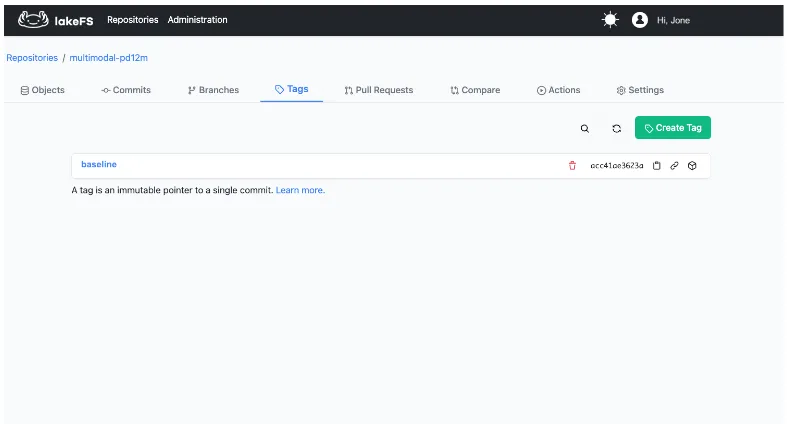
This dataset images of the baseline version are accessible via the lakefs://multimodal-pd12m/baseline/images/ path and the metadata table of that version is accessible at multimodal-pd12m.baseline.images.image_metadata.
Both the images and the Iceberg metadata table now live under the same repository, ready for multimodal ML.
Querying your Iceberg Tables with Dremio
lakeFS provides Git-like controls to organize and version your data lake, while Apache Iceberg brings transactional consistency and rich table management features. But querying that versioned data at scale, especially when combining structured and unstructured assets, requires a high-performance engine that understands the Iceberg table format.
That’s where Dremio fits in.
Dremio is a lakehouse engine designed to work directly on object storage without data movement. It natively understands Apache Iceberg, supports REST-based catalogs, and provides powerful capabilities like:
- Semantic modeling: Define views, document datasets, and enforce access control across teams.
- Accelerated queries: Automatically manage performance with reflections—Dremio’s version of transparent, auto-updating materializations.
- Open access: Query Iceberg tables and external data sources using SQL, BI tools, or Python notebooks, all from one interface.
- AI Functions: Dremio has recently introduced AI functions that’ll let you leverage your unstructured assets managed by lakeFS alongside your structured data in SQL queries. (For example: converting a PDF of hand-filled forms into rows in an iceberg table)
- Agentic Analytics: While Dremio can connect to your favorite BI tool, Dremio has an Integrated AI Agent and MCP server, allowing you to query the data and generate visualizations with Natural Language
Dremio doesn’t just run queries. It unifies the data lake, speeds up results, and lets you manage data context and governance from a single platform.
Connecting Dremio to lakeFS
To connect Dremio to a lakeFS-managed repository including Iceberg tables, you’ll use the Iceberg REST Catalog interface. This allows Dremio to interact with lakeFS repositories via a version-aware API, while keeping lakeFS out of the data path entirely. In other words, Dremio reads and writes data files directly from your object store (like S3), while lakeFS tracks snapshot metadata in the background.
Here’s how to set it up:
- In Dremio, go to the Datasets page and click the + to add a new source.
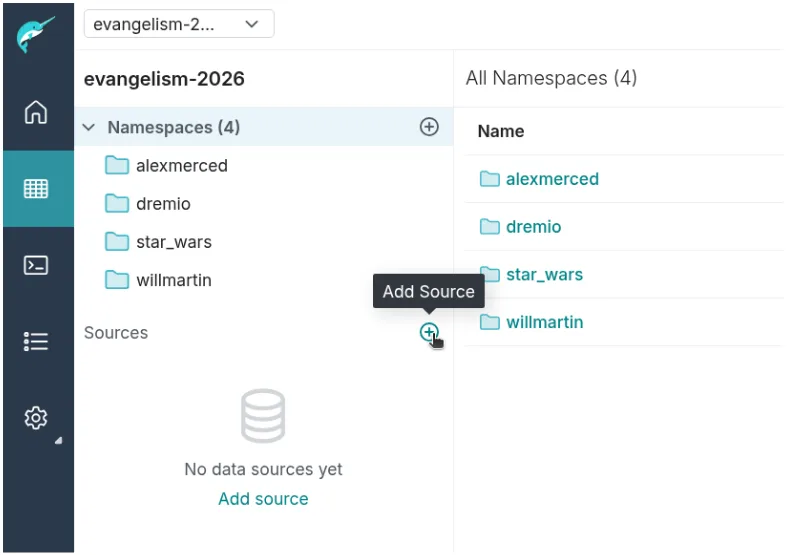
- Under Lakehouse Catalogs, choose Iceberg REST Catalog Source.
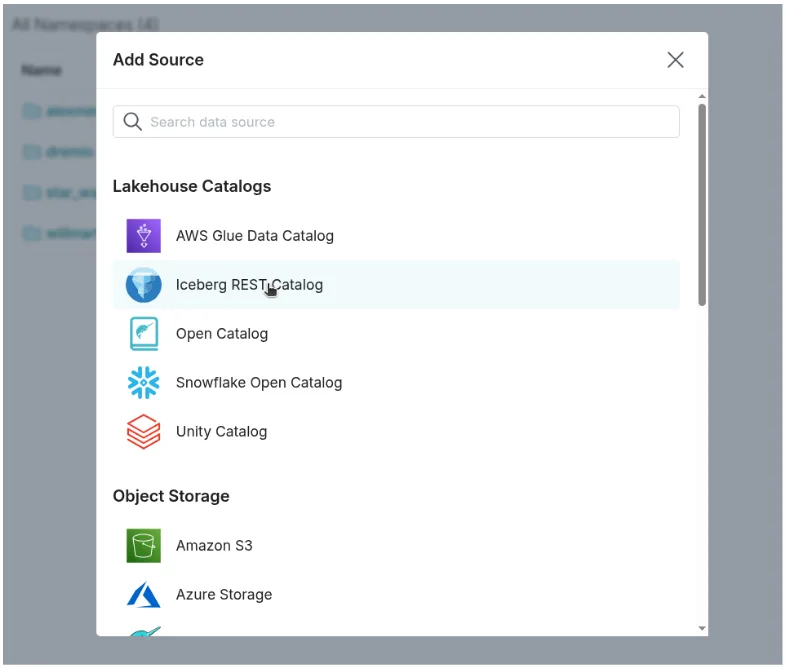
- Provide:
- A name (e.g. lakefs-catalog)
- REST endpoint URL (e.g. https://lakefs.example.com/iceberg/api)
- Uncheck Use vended credentials
- A name (e.g. lakefs-catalog)

- In Advanced Options → Catalog Properties, add the following key-value pairs:
| Key | Value |
| oauth2-server-uri | https://lakefs.example.com/iceberg/api/v1/oauth/tokens |
| credential | <lakefs_access_key>:<lakefs_secret_key> |
| fs.s3a.aws.credentials.provider | org.apache.hadoop.fs.s3a.SimpleAWSCredentialsProvider |
| fs.s3a.access.key | <your_aws_access_key> |
| fs.s3a.secret.key | <your_aws_secret_key> |
| dremio.s3.list.all.buckets | false |
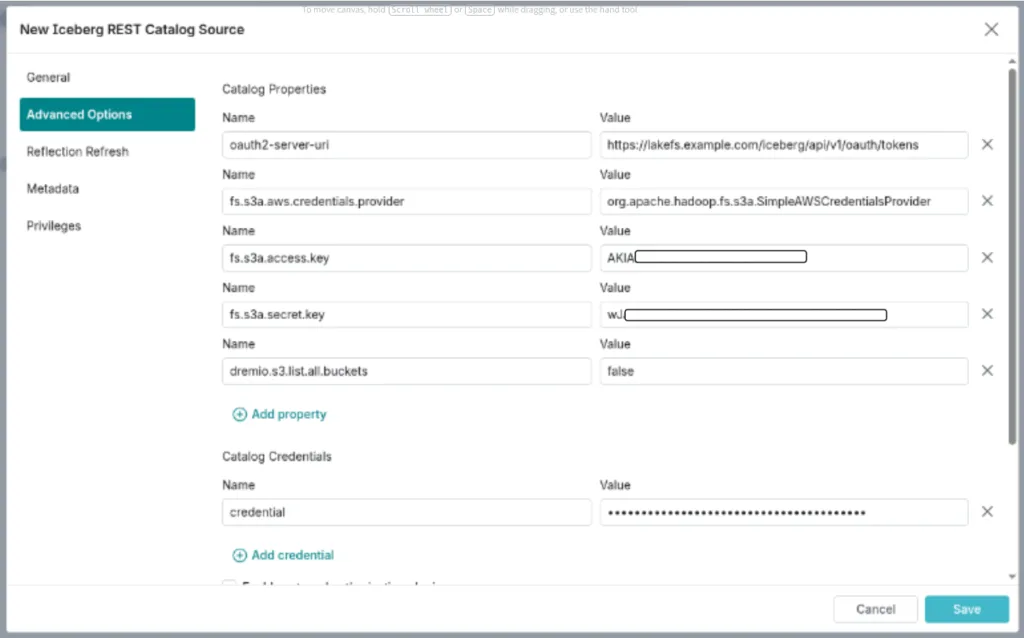
- Click Save to finalize the connection.
Once connected, you can reference Iceberg tables in Dremio using the following format:
SELECT * FROM lakefs-catalog.multimodal-pd12m.baseline.images.image_metadata;
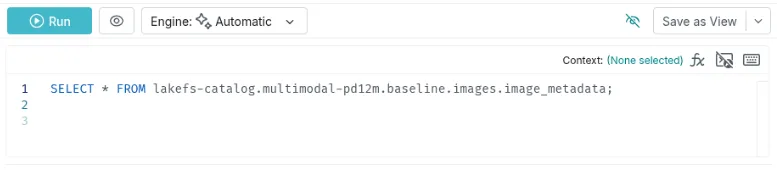
This query targets the baseline tag of the multimodal-pd12m lakeFS repository, ensuring that results reflect the exact snapshot in that tag.
With this setup, every query is anchored to a specific lakeFS version. Whether you're experimenting on a new branch or auditing a tagged historical snapshot, Dremio makes it easy to explore and analyze your Iceberg data without ambiguity or drift.
Querying Multimodal Snapshots
With Dremio connected to the lakeFS REST Catalog, querying a consistent snapshot of your data becomes seamless. This is especially useful in multimodal workflows where different data types, like Iceberg tables for metadata and object files for images or logs,need to stay in sync.
Every query you run in Dremio is tied to a specific reference in lakeFS. If you're on the ingest branch of the multimodal-pd12m repository, Dremio queries will reflect exactly what’s committed in that branch. This guarantees reproducibility and enables safe, collaborative experimentation.
You can join and analyze datasets as you normally would:
SELECT img.image_id, meta.category, meta.timestamp FROM lakefs-catalog.multimodal-pd12m.ingest.pd12m.images AS img JOIN lakefs-catalog.multimodal-pd12m.ingest.pd12m.metadata AS meta ON img.image_id = meta.image_id

Here, images could point to file-based objects imported via lakeFS, while metadata is an Iceberg table registered in the same branch. Dremio’s Iceberg support and columnar execution engine handle the join efficiently, even across data types.
Branching for Experiments
One of the most powerful workflows enabled by lakeFS is the ability to create isolated branches for experimentation. Dremio fully supports this by letting you query any lakeFS branch as if it were its own versioned catalog.
Want to test a new transformation on your data? Start by creating a new branch:
lakefs branch create experiment-01 --source main --repo multimodal-pd12m
Then, write data or register tables to that branch. In Dremio, just switch the reference in your SQL:
SELECT * FROM lakefs-catalog.multimodal-pd12m.experiment-01.pd12m.metadata;
This workflow mirrors Git branching, but for data. You can run pipelines, validate schema changes, or tune preprocessing logic without affecting production. And because the branching is zero-copy, you aren’t duplicating storage, just isolating metadata references.
Merge and Promotion to Production
Once your work on a feature branch is validated, whether through manual checks, queries in Dremio, or automated tests, you can safely merge it into your main branch using lakeFS:
lakefs merge experiment-01 --into main --repo multimodal-pd12m
Since Dremio queries are version-aware, users consuming data from the main branch will instantly see the updated snapshot on their next query. There’s no need to restart services or refresh metadata manually.
This atomic workflow means your AI-ready data pipeline can match the maturity of your CI/CD pipeline, enabling safe, fast iteration with a clear path from staging to production.
Why This Matters
Combining lakeFS and Dremio delivers a powerful pattern for managing multimodal data:
- Reproducibility: Every query is tied to a specific data snapshot.
- Safety: Branching and merging let teams experiment without risk.
- Performance: Dremio accelerates Iceberg queries using intelligent caching and reflections.
- Simplicity: One platform to query structured tables, unstructured files, and external sources—without data duplication.
Multimodal data management doesn’t have to be complex or fragile. With lakeFS providing versioning, Iceberg handling structure, and Dremio powering exploration, teams get a production-grade workflow that supports fast iteration, shared context, and long-term reliability.
Using lakeFS With Dremio AI Functions
While the Iceberg REST Catalog is ideal for querying structured data, Dremio also supports direct S3-compatible access to unstructured data stored in lakeFS. This unlocks advanced AI-driven analysis for datasets like PDFs, images, and text documents—all versioned and governed through lakeFS.
To connect Dremio to lakeFS as an S3 source:
- In Dremio, add a new Data Lake source and choose Amazon S3.
- In Advanced Options, enable compatibility mode.
- Set fs.s3a.path.style.access = true and provide the lakeFS S3 gateway endpoint.
- Enter your lakeFS credentials under the General tab.
Once connected, Dremio can browse lakeFS repositories just like any S3-compatible storage. But the real magic happens when you use Dremio’s AI functions, like AI_GENERATE, AI_CLASSIFY, and AI_COMPLETE, to extract insights from files stored in those repositories.
Let’s look at an example:
WITH recipe_analysis AS (
SELECT
file['path'] AS recipe_file,
AI_GENERATE(
'gpt.4o',
('Extract recipe details', file)
WITH SCHEMA ROW(recipe_name VARCHAR, cuisine_type VARCHAR)
) AS recipe_info
FROM TABLE(LIST_FILES('@lakefs-repo/images'))
WHERE file['path'] LIKE '%.pdf'
)
SELECT
recipe_file,
recipe_info['recipe_name'] AS recipe,
recipe_info['cuisine_type'] AS cuisine
FROM recipe_analysis;In this query, we:
- List PDF files from a lakeFS repository.
- Use a prompt and file reference to extract structured metadata.
- Return structured rows with fields like recipe_name and cuisine_type.
You can go further by classifying files with AI_CLASSIFY, or creating summaries with AI_COMPLETE. Because the source data lives in a versioned lakeFS branch, every AI operation is reproducible. You’ll always know which version of a file produced each result, no guesswork.
This lets you build intelligent data pipelines where structured and unstructured data live together, stay in sync, and feed analytics, dashboards, or model training, all from SQL.
Conclusion
Managing multimodal data, structured, semi-structured, and unstructured, is no longer just a data engineering concern. It’s an AI concern. Reproducibility, governance, and speed matter at every step.
With lakeFS, you version everything: Iceberg tables, images, models, logs. With Dremio, you query and analyze it all, structured or not, at scale. Together, they bring Git-style control and interactive querying to your data lake, so you can build more intelligent, version-aware workflows without sacrificing flexibility or performance.
Want to try it for yourself?
- Start using lakeFS for free
- Get started with Dremio and connect it to your lakeFS repo in minutes
Build more intelligent data workflows with version control, scale, and speed—all without giving up the flexibility of your data lake.